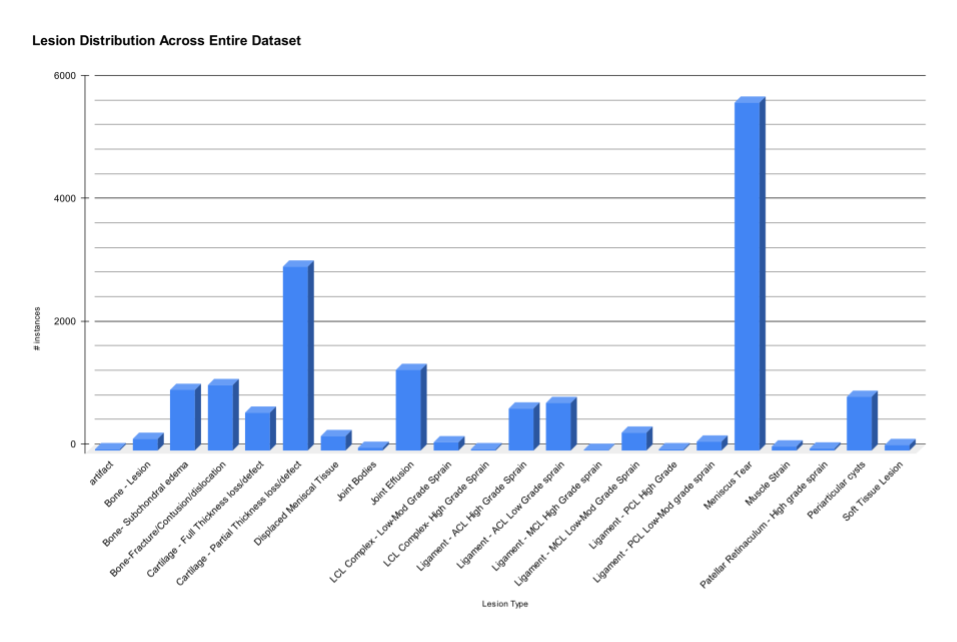
lesion detection
Detectron2 based pipeline to detect lesions present in undersampled MRI images.
To save MRI image generation time from raw MRI measurements, deep learning algorithms have emerged as a viable method for MRI reconstruction with higher rates of acceleration. Recent reconstruction problems have shown various shortcomings in current deep learning approaches, including the loss of fine image details even when employing models that score well in terms of global quality criteria. This study provided an end-to-end deep learning framework for pathology detection present in MRI images.
The proposed system accepts as input MRI images obtained using MRI reconstruction algorithms (such as VarNet), then conducts lesion identification. Using publicly available fastMRI dataset, this is the first study to establish an automated lesion identification pipeline for MRI images reconstructed using undersampled raw MRI measurements with comparable identification performance at undersampling factors upto 8.
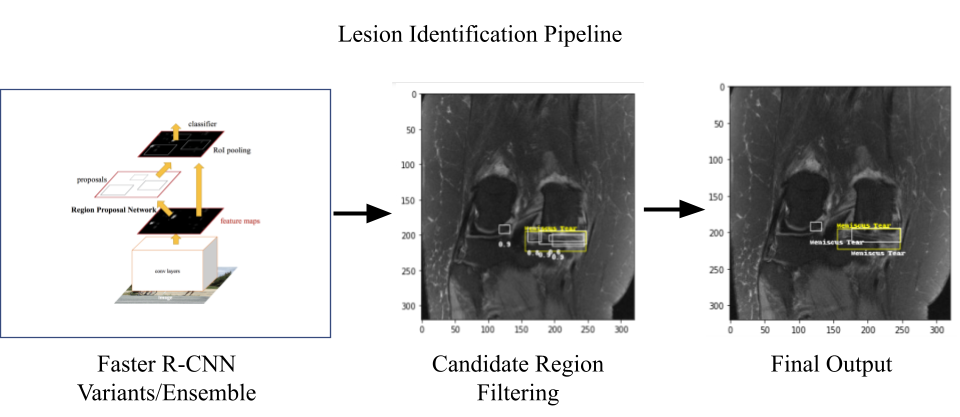
The lesion identification pipeline consisted of three different variants of faster-rcnn models present in Detectron2 library: (1) faster_rcnn_R_50_FPN_3x; (2) faster_rcnn_R_50_DC5_3x; and (3) faster_rcnn_R_50_C4. The identified lesions from all these model variants were then ensembled for final output.
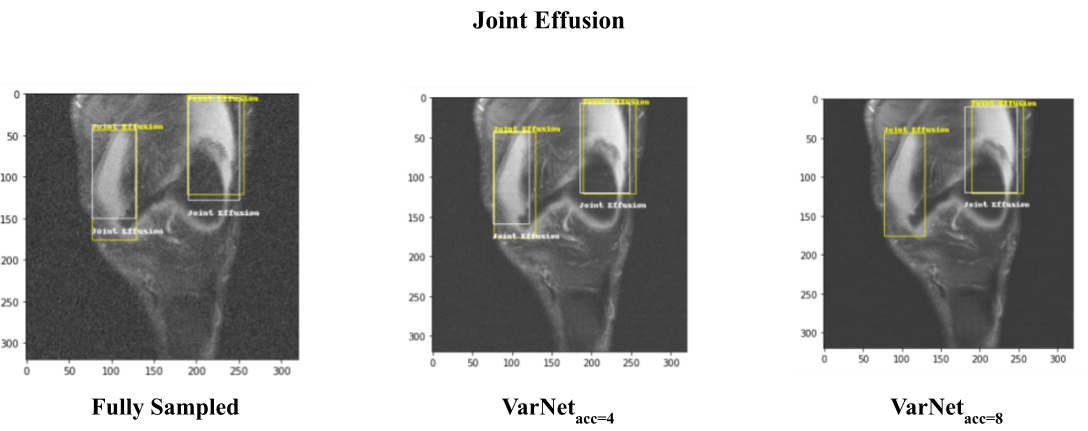
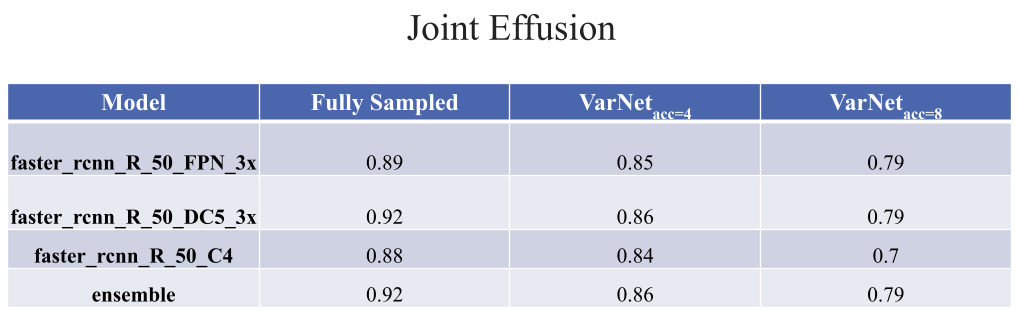
It was observed that the developed lesion identification pipeline was able to detect common and as well some rare lesions with comparable identification performance for MRI images reconstructed at various undersampling rates. More information can be found in the presentation here.